

The sequence of SMD-seq samples have been deposited in the Genome Sequence Archive ( ) with BioProject accession number PRJCA010011.įunding: Supported by National Natural Science Foundation of China Grant 21927802 (Y.H.), 22050002 (Y.H.) and T2188102 (Y.H.), Beijing Municipal Science and Technology Commission Grant Z201100005320016 (Y.H.), and Beijing Advanced Innovation Center for Genomics, also by CAMS Innovation Fund for Medical Sciences (2019-I2M-5-038). This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.ĭata Availability: Data that underlie the quantitative observations summarized in the figures and results are reported in the Supporting Information supplementary data files. Received: Accepted: JPublished: July 1, 2022Ĭopyright: © 2022 Chen et al. Fernandez-Valverde, Centro de Investigacion y de Estudios Avanzados del Instituto Politecnico Nacional, MEXICO

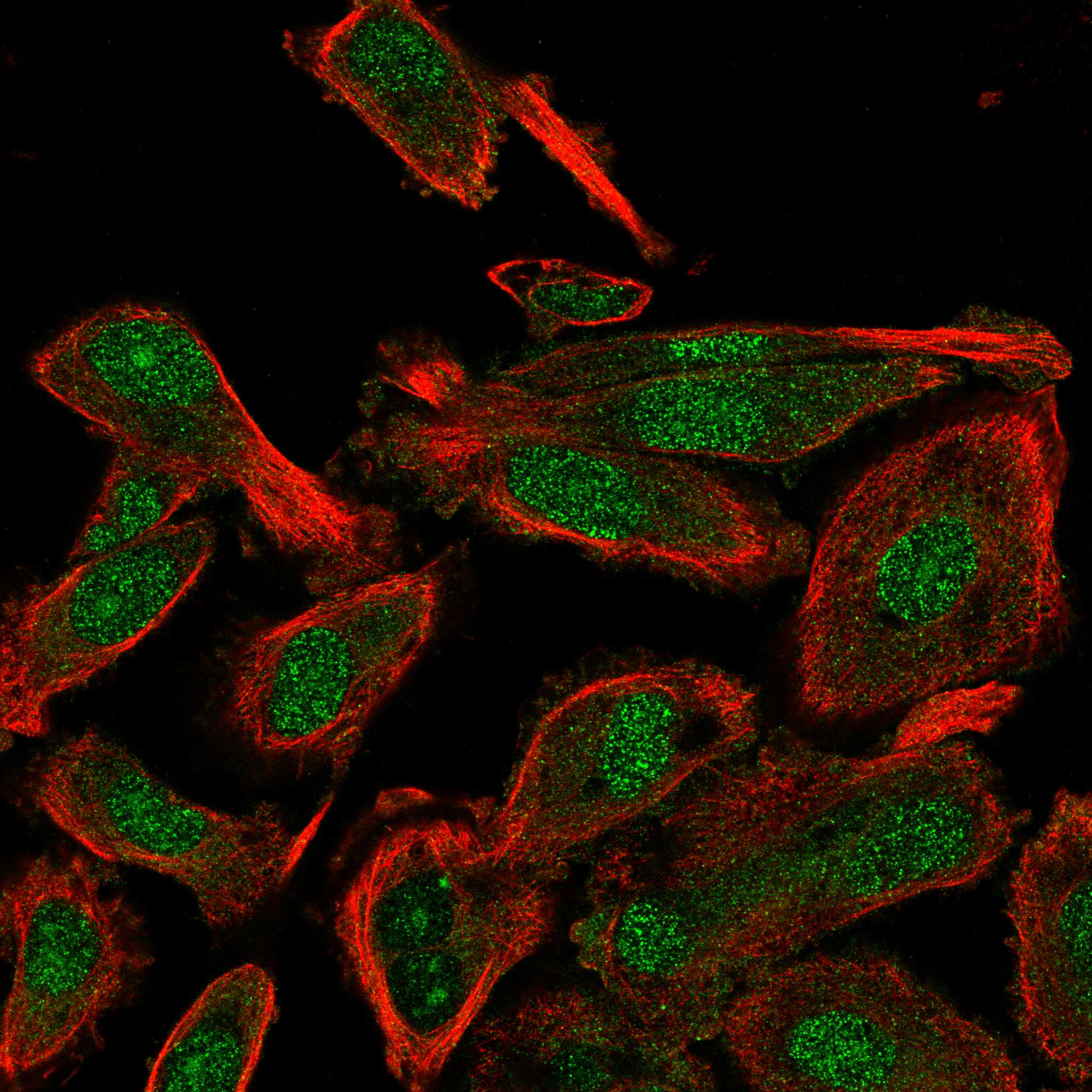
PLoS Biol 20(7):Īcademic Editor: Selene L. Our approach provides complementary insights in tumorigenesis and offers an integrative tool for macroscale cancer tissues with spatial multiomics assessments.Ĭitation: Chen T, Cao C, Zhang J, Streets A, Li T, Huang Y (2022) Histologically resolved multiomics enables precise molecular profiling of human intratumor heterogeneity. We demonstrate the capabilities of this technique by mapping gene expression and copy number alterations to histologically defined regions in human oral squamous cell carcinoma (OSCC). These microtissue samples are then processed for DNA and RNA sequencing to identify unique genetic profiles that correspond to distinct anatomical regions. This approach leverages stimulated Raman scattering microscopy to provide chemical contrast that reveals histological tissue architecture, allowing for high-resolution in situ laser microdissection of regions of interests. Here, we present a technique that combines label-free histology with spatially resolved multiomics in unfixed and unstained tissue sections. There is a growing need for tools that can be used to explore the relationship between histological features, gene expression patterns, and spatially correlated genomic alterations in healthy and diseased tissue samples. However, it can be challenging to fully characterize the molecular profile of cells in a tissue with high spatial resolution because microscopy has limited ability to extract comprehensive genomic information, and the spatial resolution of genomic techniques tends to be limited by dissection. For example, intratumor heterogeneity and the distribution of transcriptional and genetic events in single cells drive the genesis and development of cancer. Both the composition of cell types and their spatial distribution in a tissue play a critical role in cellular function, organ development, and disease progression.


 0 kommentar(er)
0 kommentar(er)
